Scalable Genomic Processing: 1,000x Speed Improvement with Distributed Computing
Built a distributed computing framework using Apache Spark for an immuno-sequencing startup, achieving 1,000x+ processing speed improvement on terabyte-scale genomic datasets with interactive visualization.
The Client
The immuno-sequencing startup spun out of a premier academic institution with a focused mission: accelerate genomic research by making large-scale immuno-sequence analysis accessible and fast. Their work centered on analyzing immune system repertoires—the billions of unique receptor sequences that define how an organism fights disease. This analysis is fundamental to vaccine development, cancer immunotherapy, and autoimmune disease research.
The founding team brought deep domain expertise in computational biology and immunology, with early research generating promising intellectual property. They had secured initial funding and needed to demonstrate that their analytical approach could scale from academic proof-of-concept to production-grade infrastructure capable of processing datasets from multiple research partners simultaneously.
The Challenge
Immuno-sequencing generates massive datasets. A single experiment can produce terabytes of raw sequence data that must be processed, aligned, clustered, and analyzed before researchers can extract meaningful biological insights. The startup’s existing analysis pipeline—built on single-machine Python scripts during the academic phase—could process a single dataset in days. At that rate, supporting multiple research collaborations was impossible.
The processing bottleneck was computational: sequence alignment, diversity calculations, and clonotype clustering are inherently parallelizable but were running sequentially. The team needed results in minutes, not days, to enable interactive exploration—researchers wanted to adjust parameters and re-run analyses in real time during working sessions. The solution also needed to be portable across cloud providers and on-premises servers, since different research partners had different infrastructure requirements and data governance policies.
Our Solution
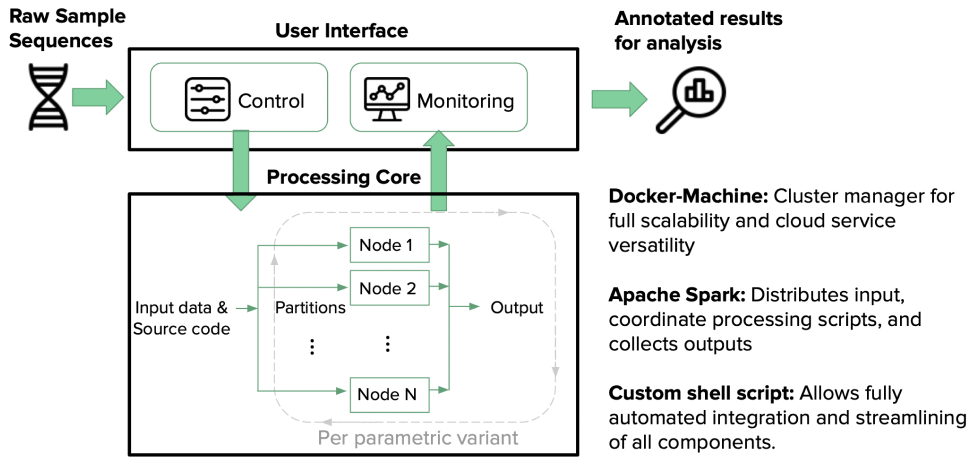
We designed and built a distributed computing framework using Apache Spark with PySpark, replacing sequential processing with massively parallel execution across compute clusters. The pipeline ingested raw immuno-sequence files, performed distributed alignment and quality filtering, computed diversity metrics and clonotype frequencies, and generated summary statistics—all leveraging Spark’s in-memory processing and automatic data partitioning.
For the interactive exploration layer, we built a Python Dash application that connected directly to processed results, giving researchers real-time visualization of repertoire diversity, clonal expansion patterns, and cross-sample comparisons. The Dash app was deployed on Heroku for immediate accessibility, with no local software installation required by research collaborators. The entire architecture was designed for cloud-service compatibility—the Spark jobs could run on AWS EMR, Google Dataproc, or on-premises Hadoop clusters without modification.
The Impact
Processing speed improved by over 1,000x—analyses that took days on a single machine completed in minutes on a distributed cluster. This transformed the research workflow from batch processing (submit a job and wait) to interactive exploration (adjust parameters, re-run, visualize immediately). Researchers could now iterate on hypotheses during a single working session instead of waiting overnight between runs.
The open-source technology stack kept infrastructure costs dramatically lower than proprietary genomic analysis platforms, making the startup’s offering financially accessible to academic research partners with limited budgets. Cloud portability meant the startup could onboard new collaborators regardless of their existing infrastructure. The platform’s performance and flexibility strengthened the startup’s position for subsequent funding rounds by demonstrating scalable, production-ready technology rather than academic prototypes.